——题目:《B.1生物导体简介》作者:马丁·摩根
日期:“16 - 17五月,2016”输出:BiocStyle::html_document: toc: true toc_depth: 2 vignette: > % \VignetteIndexEntry{B。1——Bioconductor简介}% \VignetteEngine{knitr::rmarkdown}——' ' ' {r style, echo = FALSE, results = 'asis'} options(width=100) knitr::opts_chunk$set(eval=as.logical(Sys。gettenv ("KNITR_EVAL", "TRUE")),缓存=as.logical(Sys. value)。采用“KNITR_CACHE”,“真正的”)))``` ```{r setup, echo=FALSE} suppressPackageStartupMessages({library(Biostrings) library(GenomicRanges)})高通量基因组数据的分析和理解-统计分析:大数据,技术工件,设计实验;-理解:生物学背景,可视化,可重复性-高通量-测序:RNASeq, ChIPSeq,变体,拷贝数,…-微阵列:表达,SNP,…-流式细胞仪,蛋白质组学,图像,…包。小插图。工作流程。1211软件包。也……-“注释”包——标识符图、基因模型、通路等静态数据库; e.g., [TxDb.Hsapiens.UCSC.hg19.knownGene][] - 'Experiment packages -- data sets used to illustrate software functionality, e.g., [airway][] - Discover and navigate via [biocViews][] - Package 'landing page' - Title, author / maintainer, short description, citation, installation instructions, ..., download statistics - All user-visible functions have help pages, most with runnable examples - 'Vignettes' an important feature in Bioconductor -- narrative documents illustrating how to use the package, with integrated code - 'Release' (every six months) and 'devel' branches - [Support site](https://support.bioconductor.org); [videos](https://www.youtube.com/user/bioconductor), [recent courses](//www.andersvercelli.com/help/course-materials/) Package installation and use - A package needs to be installed once, using the instructions on the landing page. Once installed, the package can be loaded into an R session ```{r require} library(GenomicRanges) ``` and the help system queried interactively, as outlined above: ```{r help, eval=FALSE} help(package="GenomicRanges") vignette(package="GenomicRanges") vignette(package="GenomicRanges", "GenomicRangesHOWTOs") ?GRanges ``` # High-throughput Sequence Analysis ## Overall Work Flow 1. Experimental design - Keep it simple, e.g., 'control' and 'treatment' groups - Replicate within treatments! 2. Wet-lab sequence preparation (figure from http://rnaseq.uoregon.edu/) 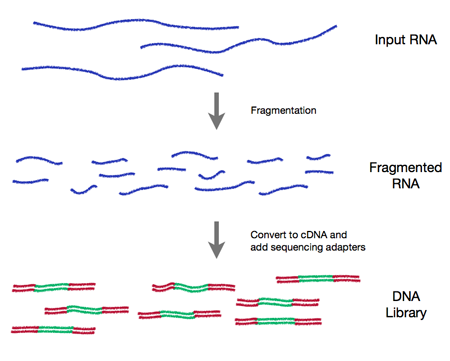 - Record covariates, including processing day -- likely 'batch effects' 3. (Illumina) Sequencing (Bentley et al., 2008, doi:10.1038/nature07517)  - Primary output: FASTQ files of short reads and their [quality scores](http://en.wikipedia.org/wiki/FASTQ_format#Encoding) 4. Alignment - Choose to match task, e.g., [Rsubread][], Bowtie2 good for ChIPseq, some forms of RNAseq; BWA, GMAP better for variant calling - Primary output: BAM files of aligned reads - More recently: [kallisto][] and similar programs that produce tables of reads aligned to transcripts 5. Reduction - e.g., RNASeq 'count table' (simple spreadsheets), DNASeq called variants (VCF files), ChIPSeq peaks (BED, WIG files) 6. Analysis - Differential expression, peak identification, ... 7. Comprehension - Biological context  # High-Throughput Sequence Data Types ## Sequencing data types Sequenced reads: FASTQ files @ERR127302.1703 HWI-EAS350_0441:1:1:1460:19184#0/1 CCTGAGTGAAGCTGATCTTGATCTACGAAGAGAGATAGATCTTGATCGTCGAGGAGATGCTGACCTTGACCT + HHGHHGHHHHHHHHDGG
> CE ?=896=: @err127302.1704 hwi - eas350_041:1:1460:16861#0/1 gcggtatgctggaaggtgctcgaatggagagcgccagcgccccggcgctgagccgccccccc>ed4 > eee > de8eeede8b ? eb <@3; ba79 ?, 881b ?@73;########################对齐读取:BAM文件-头@HD VN:1.0 SO:坐标@SQ SN:chr1 LN:249250621 @SQ SN:chr10 LN:135534747 @SQ SN:chr11 LN:135006516…@SQ号:chrY LN:59373566 @PG ID:TopHat VN:2.0.8b CL:/home/hpages/ TopHat -2.0.8b。Linux_x86_64/tophat——match -inner-dist 150——solexa-quals——max-multihits 5——no- disdant——no-mixed——covere -search——microexon-search——libraries -type fr- un——num-threads 2——output-dir tophat2_out/ERR127306 /home/hpages/ bowtib2 -2.1.0/indexes/hg19 fastq/ERR127306_1。fastq fastq / ERR127306_2。fastq -对准:ID,标志,对准和配偶ERR127306.7941162 403 chr14 19653689 3 72M = 19652348 -1413…ERR127306.22648137 145 chr14 19653692 1 72M = 19650044 -3720…ERR127306.933914 339 chr14 19653707 1 66M120N6M = 19653686 -213…-对齐:序列和质量…GAATTGATCAGTCTCATCTGAGAGTAACTTTGTACCCATCACTGATTCCTTCTGAGACTGCCTCCACTTCCC *'%%%%%#&&%''#'&%%%)&&%%$%%'%%'&*****$))$)'')'%)))&)%%%%$'%%%%&"))'')%)) ...TTGATCAGTCTCATCTGAGAGTAACTTTGTACCCATCACTGATTCCTTCTGAGACTGCCTCCACTTCCCCAG '**)****)*'*&*********('&)****&***(**')))())%)))&)))*')&***********)**** ... TGAGAGTAACTTTGTACCCATCACTGATTCCTTCTGAGACTGCCTCCACTTCCCCAGCAGCCTCTGGTTTCT '******&%)&)))&")')'')'*((******&)&'')'))$))'')&))$)**&&**************** - Alignments: Tags ... AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:0 MD:Z:72 YT:Z:UU NH:i:2 CC:Z:chr22 CP:i:16189276 HI:i:0 ... AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:0 MD:Z:72 YT:Z:UU NH:i:3 CC:Z:= CP:i:19921600 HI:i:0 ... AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:4 MD:Z:72 YT:Z:UU XS:A:+ NH:i:3 CC:Z:= CP:i:19921465 HI:i:0 ... AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:4 MD:Z:72 YT:Z:UU XS:A:+ NH:i:2 CC:Z:chr22 CP:i:16189138 HI:i:0 Called variants: VCF files - Header ##fileformat=VCFv4.2 ##fileDate=20090805 ##source=myImputationProgramV3.1 ##reference=file:///seq/references/1000GenomesPilot-NCBI36.fasta ##contig=
# #逐步=部分# #信息=
# #信息=
...# #过滤器=
# #过滤器=
...# #格式=
# #格式=
-位置#CHROM POS ID参考ALT质量过滤器…20 14370 rs6054257 G A 29 PASS…20 17330。T A 3 q10…20 1110696 rs6040355 A G,T 67 PASS…-变种信息#CHROM POS…信息…20 14370…NS = 3; DP = 14;房颤= 0.5;数据库;H2…20 17330… NS=3;DP=11;AF=0.017 ... 20 1110696 ... NS=2;DP=10;AF=0.333,0.667;AA=T;DB ... - Genotype FORMAT and samples ... POS ... FORMAT NA00001 NA00002 NA00003 ... 14370 ... GT:GQ:DP:HQ 0|0:48:1:51,51 1|0:48:8:51,51 1/1:43:5:.,. ... 17330 ... GT:GQ:DP:HQ 0|0:49:3:58,50 0|1:3:5:65,3 0/0:41:3 ... 1110696 ... GT:GQ:DP:HQ 1|2:21:6:23,27 2|1:2:0:18,2 2/2:35:4 Genome annotations: BED, WIG, GTF, etc. files. E.g., TGF: - Component coordinates 7 protein_coding gene 27221129 27224842 . - . ... ... 7 protein_coding transcript 27221134 27224835 . - . ... 7 protein_coding exon 27224055 27224835 . - . ... 7 protein_coding CDS 27224055 27224763 . - 0 ... 7 protein_coding start_codon 27224761 27224763 . - 0 ... 7 protein_coding exon 27221134 27222647 . - . ... 7 protein_coding CDS 27222418 27222647 . - 2 ... 7 protein_coding stop_codon 27222415 27222417 . - 0 ... 7 protein_coding UTR 27224764 27224835 . - . ... 7 protein_coding UTR 27221134 27222414 . - . ... - Annotations gene_id "ENSG00000005073"; gene_name "HOXA11"; gene_source "ensembl_havana"; gene_biotype "protein_coding"; ... ... transcript_id "ENST00000006015"; transcript_name "HOXA11-001"; transcript_source "ensembl_havana"; tag "CCDS"; ccds_id "CCDS5411"; ... exon_number "1"; exon_id "ENSE00001147062"; ... exon_number "1"; protein_id "ENSP00000006015"; ... exon_number "1"; ... exon_number "2"; exon_id "ENSE00002099557"; ... exon_number "2"; protein_id "ENSP00000006015"; ... exon_number "2"; ... Derived results, e.g., 'count' tables (.csv files) for RNA-seq differential expressoin. ## Major _Bioconductor_ Packages - [GenomicRanges][]: 'Ranges' to describe data and annotation; `GRanges()`, `GRangesList()` - [Biostrings][]: DNA and other sequences, `DNAStringSet()`; [BSgenome][] - [GenomicAlignments][]: Aligned reads; `GAlignemts()` and friends - [GenomicFeatures][], [AnnotationDbi][]: annotation resources, `TxDb` and `org` packages. - [SummarizedExperiment][]: coordinating experimental data - [rtracklayer][]: import BED, WIG, GTF, etc. _Bioconductor_ Objects - Example: [Biostrings][] ```{r} library(Biostrings) data(phiX174Phage) phiX174Phage letterFrequency(phiX174Phage, c("A", "C", "G", "T")) letterFrequency(phiX174Phage, "GC", as.prob=TRUE) ``` - Represent complicated data types - Foster interoperability - S4 object system - Introspection: `methods()`, `getClass()`, `selectMethod()` - 'accessors' and other documented functions / methods for manipulation, rather than direct access to the object structure - Interactive help - `method?"substr,
" '查看' substr '方法的所有帮助页,例如'方法?"letterFrequency,XStringSet" '查找' letterFrequency '泛型应用于对象(派生自)' XStringSet '类的文档。——“类?D
'来帮助那些名字以'D'开头的类(例如,用' class?DNAStringSet '找到DNAStringSet类的帮助页)。[biocViews]: //www.andersvercelli.com/packages/ [AnnotationDbi]: //www.andersvercelli.com/packages/AnnotationDbi [AnnotationHub]: //www.andersvercelli.com/packages/AnnotationHub [BSgenome.Hsapiens.UCSC.]hg19]: //www.andersvercelli.com/packages/BSgenome.Hsapiens.UCSC.hg19 [BSgenome]: //www.andersvercelli.com/packages/BSgenome [BiocParallel]: //www.andersvercelli.com/packages/BiocParallel [Biostrings]: //www.andersvercelli.com/packages/Biostrings [CNTools]: //www.andersvercelli.com/packages/CNTools [ChIPQC]: //www.andersvercelli.com/packages/ChIPQC [ChIPseeker]: //www.andersvercelli.com/packages/ChIPseeker [DESeq2]: //www.andersvercelli.com/packages/DESeq2 [DiffBind]://www.andersvercelli.com/packages/DiffBind [genome alignments]: //www.andersvercelli.com/packages/GenomicAlignments [genome features]: //www.andersvercelli.com/packages/GenomicFeatures [GenomicFiles]: //www.andersvercelli.com/packages/GenomicFiles [GenomicRanges]: //www.andersvercelli.com/packages/GenomicRanges [Gviz]: //www.andersvercelli.com/packages/Gviz [Homo.]sapiens]: //www.andersvercelli.com/packages/Homo.sapiens [IRanges]: //www.andersvercelli.com/packages/IRanges [KEGGREST]: //www.andersvercelli.com/packages/KEGGREST [OmicCircos]: //www.andersvercelli.com/packages/OmicCircos [PSICQUIC]: //www.andersvercelli.com/packages/PSICQUIC [Rsamtools]: //www.andersvercelli.com/packages/Rsamtools [Rsubread]: //www.andersvercelli.com/packages/Rsubread [ShortRead]: //www.andersvercelli.com/packages/ShortRead [SomaticSignatures]://www.andersvercelli.com/packages/SomaticSignatures [summary izeexperiment]: //www.andersvercelli.com/packages/SummarizedExperiment [TxDb.Hsapiens.UCSC.hg19.]knownGene]: //www.andersvercelli.com/packages/TxDb.Hsapiens.UCSC.hg19.knownGene [VariantAnnotation]: //www.andersvercelli.com/packages/VariantAnnotation [VariantFiltering]: //www.andersvercelli.com/packages/VariantFiltering [VariantTools]: //www.andersvercelli.com/packages/VariantTools [airway]: //www.andersvercelli.com/packages/airway [biomaRt]: //www.andersvercelli.com/packages/biomaRt [cn. cn.][生物电导]:https://生物电导.org/packages/csaw [csaw]: https://生物电导.org/packages/edger [epivizr]: https://生物电导.org/packages/epivizr [ggbio]: https://生物电导.org/packages/h5vc [limma]: https://生物电导.org/packages/metagenomeseq [org.Hs.eg.db]:://www.andersvercelli.com/packages/org.Hs.eg.db [org.Sc.sgd.db]: //www.andersvercelli.com/packages/org.Sc.sgd.db [phyloseq]: //www.andersvercelli.com/packages/phyloseq [rtracklayer]: //www.andersvercelli.com/packages/rtracklayer [snpStats]: //www.andersvercelli.com/packages/snpStats [dplyr]: https://cran.r-project.org/package=dplyr [data. data. db]: //www.andersvercelli.com/packages/org.Sc.sgd.db [phyloseq]: //www.andersvercelli.com/packages/phyloseq [rtracklayer]: //www.andersvercelli.com/packages/rtracklayertable]: https://cran.r-project.org/package=data.table [Rcpp]: https://cran.r-project.org/package=Rcpp [kallisto]: https://pachterlab.github.io/kallisto